Covid-19 Preprint Counts Are Inflated
Analysis suggests common count of Covid-19 preprints may be two times too high
An analysis conducted by “The Geyser” has found that one commonly used estimate of the number of Covid-19 preprints may be more than twice as high as it should be.
The analysis examined the most current version of the open data from Nicholas Fraser and Bianca Kramer (posted December 16), which currently estimates ~35,000 Covid-19 preprints.
The problems identified appear in multiple generations of the data, and seem structural.
These data have been quoted, used in charts, or referenced multiple times since early 2020:
- earlier and recent reporting in Nature
- a May version of a preprint on bioRxiv by Fraser et al
- a September revision of this preprint on bioRxiv
- an article about medical preprints in JAMA
- an opinion in Frontiers in Public Health
- a Letter to the Editor in Clinical Orthopaedics and Related Research (CORR)
- an article in Fast Company
- a post on “The Scholarly Kitchen”
- a “Weekend Reads” compilation on Retraction Watch
- research article coverage on INFODocket
- the “Immunobiologists” blog — in German and English
- a feature on Enago Academy
- an article in The Conversation
- a post on the blog of the Innovation Genomics Institute
- numerous social media posts (including some with confident and colorful charts)
- as the basis for a preprint on medRxiv claiming to model the growth of Covid-19 preprints
I originally detected some potential problems in the Fraser/Kramer data while writing a December 17th post about a Nature article. The article sought to distill publishing activity around Covid-19 into seven charts. In analyzing the Fraser/Kramer data used in one Nature chart, I easily detected duplicates among the listings. This suggested a high rate of duplication, and led me to conduct a more systematic analysis.
To me, measuring preprinting means measuring unique reports, not documenting the ability of authors or automation to post the same report again and again. For their part, Fraser/Kramer seem to walk both sides of this question, noting in their data repository:
. . . some preprints are deposited to multiple preprint repositories — in these cases all preprint records are included.
While implying duplication may be a slight issue, they also note efforts to prevent the inclusion of some duplicate entries, when these can be easily excluded.
I think the general assumption that each preprint is a unique report has informed most reporting and citation of these data, with double-posting not suspected, thought not to happen, or thought to be a relatively rare event. It certainly hasn’t factored into charts, discussions, or interpretations of the data, including those by Fraser/Kramer in their preprints.
Given the amount of detritus in the data, and the way it was binned, finding duplicates takes some work — but they are there, and in some quantity. Add to this the problem of counting an index as preprints, and you get a compounding effect.
At the end of the day, there are two main problems with the source data — inclusion of an irrelevant data source, and a failure to identify and correct for a large number of duplicates.

Problem #1: An Irrelevant Data Source
The first step toward a better estimate involves eliminating entries from a source that doesn’t belong. Fraser/Kramer’s Covid-19 preprint data counts entries from a bibliographic index (RePEc) as preprints, inflating the overall data set by 3,755 entries, or more than 10% of the ~35,000 preprints estimated by the authors.
RePEc (Research Papers in Economics) is a bibliographic index of articles, including many preprints. As their home page describes it:
The heart of the project is a decentralized bibliographic database of working papers, journal articles, books, books chapters, and software components, all maintained by volunteers. The collected data are then used in various services that serve the collected metadata to users or enhance it.
Glancing at at the site, it seemed clear it isn’t a preprint server — the wording is right at the top of the home page, after all — but to make sure, I emailed the administrators. Christian Zimmerman of the St. Louis Federal Reserve Bank responded right away, writing:
Indeed, we do not host, we index.
Problem #2: Failure to Remove Duplicates
The next step was to see if preprints with identical titles existed across servers. Because the raw data was sorted by platform, and a lot of “data dust” existed among the titles, duplicates weren’t going to jump out unless you looked for them.
After removing some data artifacts, the first focus was on preprints with titles matching each other 100% (or word-for-word). This pass found 1,864 preprints with at least two definite duplicate title entries — preprints with the same title and two or more postings. While most were only duplicates, groups of three or more weren’t uncommon. One preprint in this group has been posted on 11 different preprint servers.
Eliminating these duplicates reduced the number of unique preprints to 896 — a decrease of 52%. This calculates out to an average of preprints being posted more than two times each. The major servers showing duplication between preprints with identical titles are SSRN, medRxiv, and Research Square.
So much for cultural norms discouraging double-publication . . .
When it comes to duplicates, it was — and wasn’t — surprising to find preprints posted two times or more in such large numbers. After all, authors are desperate for attention and funding, Covid-19 looked like a great opportunity to pivot into the spotlight, and a sense of panic was in the air — all of which led to predictable behaviors in a system under pressure. Among peer-reviewed journals, one Covid-19 paper was published three times, with three different titles. But the implications of unreviewed reports promulgating across servers and gaining identifiers and brands bear examination — along with the implications of unreviewed open data being promulgated without question, of course. We have become self-flooding.
Many preprints have been posted even more profligately than average. Instances of three or more postings per preprint aren’t unusual in the matched data. As mentioned above, one preprint has been posted under the same title (“COVID-19 Outbreak Prediction with Machine Learning”) on 11 different preprint servers — Preprints.org, medRxiv, SSRN, Research Square, OSF Preprints, EdArXiv (OSF), engrXiv (OSF), Frenxiv (OSF), PsyArXiv (OSF), SocArXiv (OSF), and EcoEvoRxiv (OSF). According to the data, it was posted first on Preprints.org on April 19, then on medRxiv and SSRN on April 22, on Research Square on May 9, and then on the OSF properties between October 5-10. The authorship and affiliations are identical on all versions.
Because the Fraser/Kramer data is messy, another confidence band of title matching was used to detect duplication — an 80-99% title match estimate.
Among these preprints — which visual inspection suggests have a matching accuracy range closer to 95-99%, as diacriticals and other irrelevant tags account for most of the differences — there were 1,772 posting events remaining after the RePEc index entries were removed. Eliminating likely duplicates from this set left 839 unique preprints, reducing the count by 53.3%, which is about what we found in the word-for-word band. The main servers in this group are medRxiv, SSRN, arXiv, Research Square, and JMIR.
It’s worth recalling that the Fraser/Kramer data and similar data from Dimensions appeared to correlate highly at a top level in one of the Nature charts:
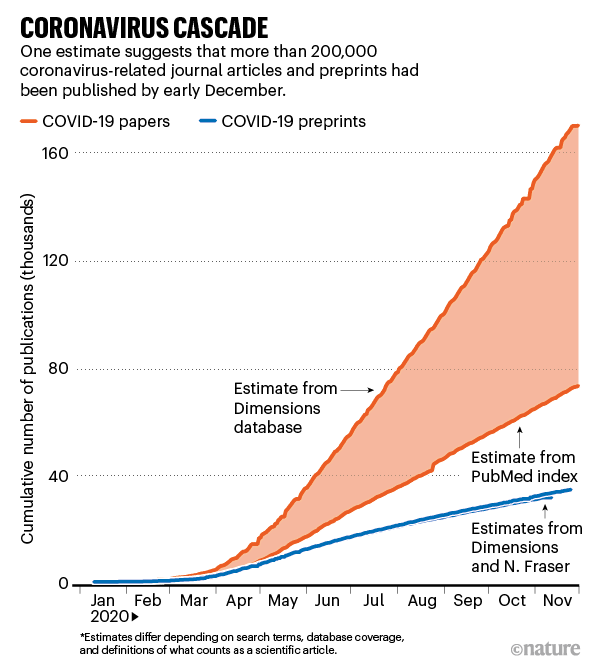
Duplicate or miscategorized preprints might exist in other comparable data sets, as well.
There may be less duplication in lower-confidence matching bands. However, this isn’t certain. One pass geared to 60-79% matching of titles revealed that the title data gathered by Fraser/Kramer are littered with the aforementioned stray code and diacritical artifacts, which confound matching — yet, visual inspection suggests there are a lot of matches remaining to be found. There is also the possibility that authors may change titles of preprints, depositing the same report on various servers under slightly different titles.
Duplicates on the Same Preprint Server
One unusual finding in the analysis of exact match titles was the number of times preprints with the same title were recorded in the data as repeatedly deposited on the same preprint server — 87 times (9.7% of the corrected total).
In the case of SSRN (22 duplicates), the duplicate records in the Fraser/Kramer data are a result of versioning, as SSRN assigns different DOIs to different versions of a preprint. This double-counting of versions inflates the counts slightly. Versioning is a form of duplication Fraser/Kramer stated they wanted to eliminate.
Research Square had 52 duplicate entries, the most by far. Others with instances of duplicate title preprints were OSF Preprints (5), Authorea (2), ChemRxiv (2), bioRxiv (2), and Preprints.org (1).
For Research Square, I could find only one instance of versioning among the duplicate-title preprint deposits — in 51 of the 52 cases, the duplicate preprint was treated as a new, independent preprint on the platform.
In one case, a preprint (“Healthcare Workers Preparedness for COVID-19 Pandemic in The Occupied Palestinian Territory: A Cross-Sectional Survey”) was deposited on medRxiv on May 13 (10.1101/2020.05.09.20096099), and then three additional times on Research Square — once on June 9 (10.21203/rs.3.rs-30144/v1), again on October 15 (10.21203/rs.3.rs-90301/v1), and once more on November 24 (10.21203/rs.3.rs-112643/v1). Because Research Square preprints seem tied to submissions to Nature journals, the unusual behavior on this server may be due to authors submitting to multiple Nature journals over time, and depositing a preprint each time.
In a puzzling instance of duplicate preprints and a single peer-reviewed publication, there is this — two preprints on bioRxiv listing slightly different author groups (a March 14th version with 10 authors, and a March 20th version with 9 authors, with some substititions occurring) leading to a paper published October 30th in Pharmacology Research & Perspectives listing the 10 authors from the first version, and with both preprints pointing to the same published version without pointing at one another.
The preprint world is messy.
Another minor source of inflated preprint counts comes from counting open peer-reviews of preprints as preprints themselves. These mainly emanate from platforms like JMIR and Copernicus GmbH.
Conclusion
Overall, adding up the two high-confidence sets, we get this:
- A claim of ~35,000 preprints is reduced by more than 10% once an irrelevant data source (the RePEc index) is eliminated, dropping the non-deduped, corrected estimate to ~31,000.
- A reduction of 52% and 53% between two bands of 3,636 claimed preprints, leaving a total of 1,735 unique reports.
- If these corrections were applied to the overall data set, the total number of Covid-19 preprints could be as low as ~14,800, rather than the ~35,000 number often portrayed or quoted. This would represent a reduction of 58% in the gross estimate, leaving 42% of the original estimate.
Fraser and Kramer’s data set about Covid-19 preprints seems flawed in at least two ways — it includes things it shouldn’t, and it fails to reduce the entities to their basic elements.
Getting to some sort of ground truth with data is complicated and time-intensive, especially when the data are this messy and confounded — or the analysts make blunders. When making charts out of preliminary data, it seems prudent to have explored, tested, or questioned it, or such charts can cement wrong impressions. GIGO will always haunt us. This goes double if we start amplifying information before satisfying ourselves that it’s worth amplifying.
That said, the data are potentially useful once cleaned up and analyzed with narrower inquiries and more careful and thorough QA.
But I’m still a little worried these data are even murkier than I’ve realized . . .
Subscriber Bonus
An accompanying analysis of which preprint servers have been upstream of others for Covid-19 preprints — that is, where authors tended to first deposit and move next — as well as an analysis of time between duplicate postings, is available to subscribers by clicking the button below.
Thanks to EA, for his help constructing the queries around the title matching bands.